Parallel corpora can be built from:
non-aligned texts
in common document formats
aligned texts
in a tabular format, e.g. .xlsx or .tmx
vertical file 1:1
expert users with 1:1 mapping
vertical file M:N
expert users with M:N mapping
Use the corresponding tab below for more information.
Parallel corpus from non-aligned documents
Parallel corpora can be built from non-aligned texts in common document formats, e.g. from two PDFs where one is the translation of the other. The supported formats are: .doc, .docx, .htm, .html, .pdf, .txt
Only 2 languages are supported when building a corpus using this method.
Automatic alignment
After uploading, the documents will be converted into plain text and aligned automatically at the sentence level and processed into a corpus. The whole process does not require intervention by the user.
For best results
- The documents must be translations of one another. (Not random texts about a similar topic.)
- Documents containing only text in one column produce best result.
- Documents with complex design such as advertisements, promotional leaflets or posters may be impossible to align and produce a poor result.
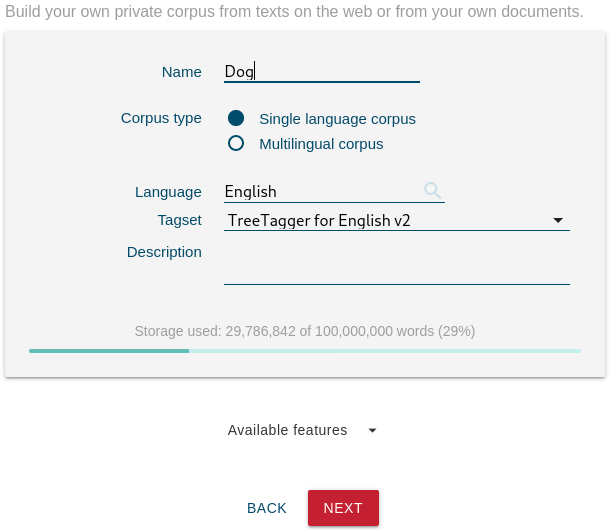
How to build a parallel corpus from documents
- go to DASHBOARD dashboard
- click NEW CORPUS
- type a name and click Multilingual corpus, click NEXT
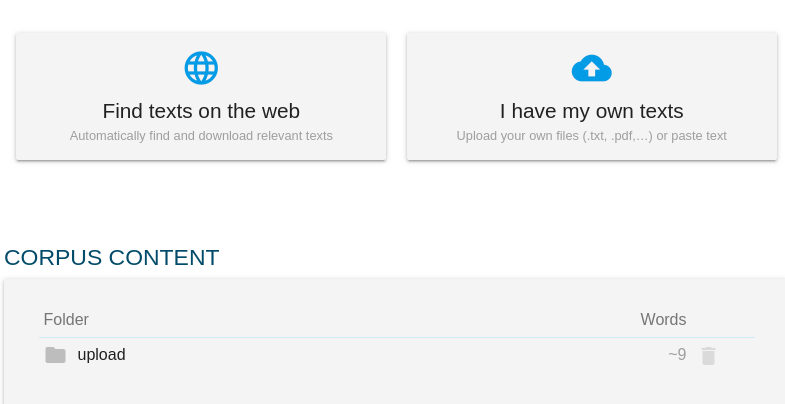
- click NON-ALIGNED DOCUMENTS
- select the languages and type the names of the corpora, for simplicity, use the same name for both languages
- upload the documents, multiple documents are supported but they must be uploaded in the same order in both languages
- click NEXT and wait for the corpus to be processed and compiled
Correcting alignment errors
Alignment errors cannot be corrected in Sketch Engine. If they are too many, they have to be corrected outside Sketch Engine. Download the corpus in one of the available formats, e.g. XLSX and use Excel or Google Sheets to correct the alignment.
Analyse the corpus
Learn to work with the parallel corpus on our YouTube channel or in this guide.
Adding more data
Additional data cannot be added using the above procedure. Instead, built a new corpus, download it and add it to the first corpus using the same procedure as the one used for aligned data.